Advanced Science News piece:
Artificial cells with specialized internal chemistries could revolutionize how we approach precision medicine.

https://www.advancedsciencenews.com/artificial-cells-valuable-niche-in-medicine/
Advanced Science News piece:
Artificial cells with specialized internal chemistries could revolutionize how we approach precision medicine.

https://www.advancedsciencenews.com/artificial-cells-valuable-niche-in-medicine/
Kate participates in The Dissenter webinar “Synthetic Cells, Cell Evolution, the Origin of Life, and Astrobiology”.
Kate was a guest of EBRC In Translation podcast, discussing science, safety and security of synthetic cell engineering ebrcintranslation.buzzsprout.com/1581817/9465178-9-building-cells-and-synbio-in-space-w-kate-adamala


.
|
|
Build-a-Cell: Engineering a Synthetic Cell Community |
|
|
Synthetic Cells in Biomedical Applications |
Our simple, 3D printed liposome formation device is now available to all non-profit users: device, protocols and tutorials
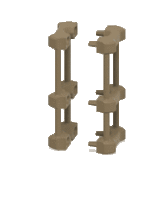
CRS News published our text about synthetic cells in drug delivery applications.
Synthetic cells: happy middle between liposome drug delivery and engineered natural cells
