Building semi-synthetic minimal cells
 Synthetic minimal cells are programmable liposomal bioreactors capable of expressing proteins from DNA (or RNA) genome. The programmability of synthetic cell metabolism and lack of underlying endogenous live cell pathways make it ideal tool for studying natural cell pathways and circuits, and for prototyping new biological systems.
Synthetic minimal cells are programmable liposomal bioreactors capable of expressing proteins from DNA (or RNA) genome. The programmability of synthetic cell metabolism and lack of underlying endogenous live cell pathways make it ideal tool for studying natural cell pathways and circuits, and for prototyping new biological systems.
We are combining top-down and bottom-up approaches to synthetic biology; using tools of protein engineering and molecular biology, together with novel synthetic cell technologies, to understand and modulate biological processes.
Synthetic minimal cells can help us studying basic properties of life, understand the origin of life on Earth, and give guidance looking for life elsewhere in the Universe. We aim at building models for studying diseases, for creating novel therapies and for making custom biofactories.
We apply engineering principles to biology: we build life, or grow machines.
This talk gives broad overview of our lab’s work.
The top-down approach involves building on the biological ensemble, modifying existing cellular pathways to explore and control biology.
The bottom-up approach involves building chemical systems capable of mimicking cellular processes – the synthetic minimal cells, that mimic properties of live cells. Those synthetic cells allow us studying and engineering complex biological pathways, building new biological functionalities, make better drugs and novel therapy strategies, and tracing the evolutionary history of life in model protocells.
Synthetic cells are liposomal bioreactors that can mimic natural biology, as well as create new functionalities from biochemical parts. Building cells from scratch, we study the origin and early evolution of life, we study complex modern cells and explore alternative ways to do “life”. We use synthetic cell technologies to make tools for metabolic engineering, drug development, and biosensing. We explore possibilities of using synthetic cells as biocomputers, and as bioreactors for space missions.
We use techniques of cell-free protein expression, liposome encapsulation and RNA engineering, combining existing synthetic biology tools and building new ones.
Lineage-agnostic biology of synthetic cells allows us to apply engineering principles to study biological processes, with an experimental platform that we control and understand, while maintaining complexity relevant to natural life. Synthetic cells can be used as both models of complex biological processes and a chassis for desininig new biological systems.
Synthetic cells 101
TED talk on synthetic life
Kate presented concept of building synthetic minimal cells, with its biotechnological, biomedical and basic science implications, in a TEDx talk .
Selected editorials and popular audience owerviews
The effort towards building life in the lab and elucidating the origin and earliest evolution of life has always been receiving interest from broad audience.
STAT magazine published nice overview of efforts to build artificial life,  From chemicals to life: Scientists try to build cells from scratch.
From chemicals to life: Scientists try to build cells from scratch.
This great editorial in Science is intorducing new, exciting paper and highlighting interesting aspects of building synthetic cells.
Excellen editorial in Nature, describing rationale of engineering synthetic life, and the most common approaches to various problems we’re facing en route to a synthetic cell.
And local copy pdf
Kate’s article on personalised medicine using synthetic cell technologies
iBiology synthetic cell talks:

Part 1: Synthetic Cells: Building Life to Understand It

Part 2: Synthetic Cells: Approach and Applications
Some examples of the astrobiology focus: a review detailing our work on RNA replication in protocells: Angewandte Chemie International Edtion Citric Acid and the RNA World 2014, 53, 5245 – 5247, external link (local copy pdf)
 Aaron and Kate did NPR podcast on the origin of life – Brains on!
Aaron and Kate did NPR podcast on the origin of life – Brains on!
Our work on protocells have been featured in science news outlets and editorials, including:
Science Focus: Robert Service, The Life Force, external link (local copy pdf)
BioTechniques: How Cellular Life Evolved 06 Jan 2014 external link (local copy pdf)
Science News: To cook up life, just add citrate 185(1):15 external link (local copy pdf)
Science Daily: Researchers find missing component in effort to create primitive, synthetic cells, external link (local copy pdf)
Chemical & Engineering News: Lab-Made Protocells Show Hints Of Evolution, 91(21), 2013
The Panda’s Thumb: New Szostak protocell is closest approximation to origin of life and Darwinian evolution so far, external link (local copy pdf)
Studying natural biology
We use synthetic cells to reconstitute and study natural biological processes, and to prototype and validate bioengineering tools that can be later used in live natural cells.
Engineering generalized RNA-protein interactions: a toolbox for regulation and readout of gene expression
We developed and validated protein architecture which binds to single stranded RNA. Using this protein technology, we are developing tools for visualization and quantification of levels of expression of genes of interest. This tool will work by following the reconstitution of a protein probe upon interaction of sequence-specific RNA binding proteins with the mRNA of the gene of interest.
We are also aiming to edit the transcriptome of the gene of interest, selectively decreasing the level of expression of one splice variant (as opposed to cutting the DNA of the gene, which targets all splice variants indiscriminately).
This technology could potentially help in studying non-ER translation events, elucidating mechanisms of synaptic plasticity, as well as studying healthy and diseased translational profiles of genes, e.g., those involved in oncogenesis and other disease processes.
The ability to monitor and perturb RNA in living neurons – which would open up the investigation of many processes that contribute to development, plasticity, and disease progression – would benefit greatly from a method of systematically targeting unmodified RNA sequences for observation and control. The current ssRNA targeting methods are based on the RNA binding protein aptamers, like MS2 or PP7; this require the introduction of aptamer binding sites into the RNA. This work shows that it is possible to develop a ssRNA binding protein that can be engineered to target arbitrary sequences of variable length, thus eliminating the need to engineer the target sites into the RNA of interest.
Popular science description: news release, local copy pdf.
This work was done with Ed Boyden
Thank you for support: NIH and Jeremy Wertheimer Foundation.
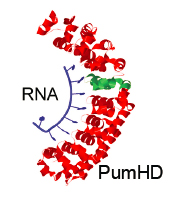
Programmable RNA-binding protein composed of repeats of a single modular unit;
Katarzyna P. Adamala*, Daniel A. Martin-Alarcon*, and Edward S. Boyden;
PNAS, 2016, 10.1073/pnas.1519368113; *equal contribution
local copy pdf
publisher website link
Biocomputing
As we are approaching the limits of technological abilities of silicone microchips, biological computation is one of the alternatives for future development of computing devices.
Due to the complex and unpredictable nature of live cell biochemistry, we use simpler synthetic minimal cells to engineer and validate biological circuits.
We engineered biocomputing platform for constructing Boolean logic gates in synthetic cell system.
TRUMPET stands for Transcriptional RNA Universal Multi-Purpose GatE PlaTform.
The platform allows constructing orthogonal universal Boolean logic gates, including layering gates to engineer complex circuits. The logic gate design tool includes sequence validation and folding tests. More at trumpet.bio
Very large-scale genetic circuit design automation
We will build liposome based synthetic minimal cell based soft-computing circuits using genetic logic gates and sequential synthetic cell fusion. The proposed research will lead to a 105 scale-up of our ability to implement information processing in cells.
Large, complex genetic circuits will be implemented in both living cells and non-living vesicles, offering extremely dense and low-power computing. The proposed research spans the scale from computing in individual cells to that performed by multicellular ecological systems.
Here, insight is taken from software engineering to make genetics programmable. Key to our approach is refactoring genetics such that units of regulation are simple and modular and thus are able to be put together by EDA software.
In other words, it is not enough to build software to model genetics, rather it also requires building the genetics that can be put together by the software. This has led to new design principles in modularity and insulation, which we extend here by utilizing the principles of multicellular computing.
The use of non-living cell free systems and synthetic cells is a potential path towards systems that incorporate physical materials from biology and semiconductors.
Thank you for support: NSF and Semiconductor Research Corporation.
This work is done in collaboration with Eduardo Sontag and Chris Voigt.
 Origins of life
Origins of life
The earliest evolution of life included a series of transition from non-living matter, through prebiotic organic synthesis and chemical evolution, towards the Last Universal Common Ancestor of all life.
Our work focuses on the immediately-pre-life stage of evolution, when chemistry became biology.
We create synthetic minimal cells that exhibit some key properties of life, without being entirely alive. Those cells express proteins inside phospholipid liposomes, using cell-free protein expression systems. Thus, represent the latest stage of prebiotic evolution, after the establishment of the Central Dogma. Those cells do not exhibit active homeostasis, but they can maintain separate internal environment, they can grow, divide and evolve. The controllability and flexibility of those minimal cells allow us studying chemical processes underlying major transitions in evolution.
In our work, we create synthetic minimal cells expressing complex genetic pathways, with membrane proteins facilitating communication with external environment. Together, this creates a comprehensive system to study the advent of cellular processes on the boundary between prebiotic and Darwinian evolution.
Thank you for support: NASA and NSF.
![]() We are part of the COOL center.
We are part of the COOL center.
The Cool Team is conducting an interdisciplinary investigation of the origins, evolution and significance of translation, which is a window to primeval events and molecular structures. We will integrate biophysical, structural, bioinformatic, biochemical, chemical and evolutionary approaches to understand biology’s “internal fossil record”. Our overriding scientific question is: How did the original and most profound mutualistic relationship in biology, of polypeptide and polynucleotide, take root and ultimately come to define and rule the biological earth? The central question will be framed by four themes: (Theme 1) Evolution of translation in vivo, (Theme 2) Evolution of translation in vitro, (Theme 3) Walking back from extant biology and (Theme 4) Origins of MolecularMutualism.
![]()
Some of this work is done as part of The Chemical Ecosystem Selection Paradigm for the Origins of Life collaboration.
The Chemical Ecosystem Selection Paradigm for the Origin of Life (CESPOoL) project will combine theoretical models of autocatalysis and surface pattern formation with multilevel selection theory to better understand the spontaneous emergence and adaptive evolution of life-like, autocatalytic chemical systems associated with mineral surfaces. Then, we will use this theory to refine implementation of a prebiotic analog of microbial ecosystem selection that has the potential to find surface-associated, life-like interacting molecular ensembles (SLIMEs) and enrich them to the point where they can be characterized and compared to biological life.
Space biology

Synthetic cells can be used as a chassis for biological and biomedical experiments under the conditions of space flight: performing reliable RNA and protein production and formation of drug particles will be crucial for establishing human presence on the Moon, Mars colony and for maintaining the health of people and animals during space flights and colonization. They can be prepared in advance and stored, lyophilised, for a long time.
We have developed synthetic cell epxerimenta payload, with a team of the RockSat-C 18 program. The payload was succesfully launched in a Terrier Orion sounding rocket from NASA Wallops in June 2018, increasing the TRL of this project to 6. This was, to our knowledge, the first time cell-free protein expression experiment was performed under spaceflight conditions.
Synthetic minimal cells have been proposed as a possible tool for biomanufacturing during long-term space missions and on extraterrestrial colonies, as well as a tool for future terraforming efforts. Synthetic minimal cells are bioreactors built specifically to mimic certain aspects of cellular processes, by reconstituting specific genetic pathways, encapsulated within a biomimic membrane. Thus, synthetic minimal cell technology combines most advantages of classical in vitro studies, with added capabilities, including spatial interactions, and interactions between many scaffolded and encapsulated proteins at once.
Traditionally, the information contained in the genetic material can travel only as far and as fast as the mass of the nucleic acids being duplicated during cell division. In the case of synthetic cells, since their autonomous reproduction is unnecessary, once a winning formula for a particular synthetic cell is discovered locally, it can simply be transmitted as digital information riding on optical fibers or radio waves at high speed (and using digital compression makes high bit rates and practically zero error rates feasible with currently available apparatus). Such information can be reduced back to synthetic cells anywhere with a synthetic cell generating apparatus ‐ be it in another country, spaceship or planet.
Our work focuses on developing mission architecture: preparation, storage and experimental conditions compatible with various flight scenarios. We are building syntetic cells to model various natural processes, to study elements of biology under the conditions of spaceflight. The primary goal of this research is to gather information about how natural terrestrial organisms behave under space flight conditions, for the benefit of designing human space exploration strategies. The other goal of this research is to help refine physicochemical boundaries for biochemistry as we know it, to aid in designing life detection systems.
All of this work is done in collaboration with Aaron Engelhart.
Metabolic engineering
Natural cells have a whole set of endogenous metabolic processes, pathways that use small molecules for suporrting the natural cell’s metabolism and growth. One of the biggest challenges in modern metabolis engineering is to integrate newly constructed pathways with the host metabolism. When trying to discover a new pathway, or improve efficiency of an existing enzyme pathway, we often run into the problem of not being able to tell whether the changes have the desired effect in the background noise of endogenous cell metabolism.
Synthetic minimal cells have no endogenous metabolism beyond what is engineered into those cells in the first place. Therefore, we can understand and control flux of small molecule substrates within the synthetic cell. This makes designing new pathways and optimizing existing ones much easier. Additionally, the short turnaround time of a synthetic cell experiment, without needing to wait for transformants to grow to test each new plasmid combination, make iterating metabolic engineering designs much faster.
Our research focuses on engineering methods for high throughput, combinatorial design of multi-gene pathways using synthetic cells as the chassis for testing and optimizing expression.
For large scale natural product synthesis, synthetic cells are mostly research and development tool, at this point it is still cheaper and easier to perform industrial scale production in a live bug. Synthetic cell screens can help identify candidate pathways, and optimize it before scaling up for industrial manufacturing.
Synthetic cell systems can be a good tool for small scale, pilot studies, or for rapid prototyping of on-demand production of small batches of biologicals or precious small molecule compounds.
Astropharmacy

During long term space missions, astronauts will encounter situations that are impossible to fully predict in advance. The isolation of the crew, for many years at a time, means that mission architecture needs to include plans for as many unpredictable situations as possible. The harsh environment of space and other planets will put high pressure on astronaut’s bodies. Providing on-demand, customizeable medical care under those conditions is very challenging.
We cannot plan for all pharmacy needs of astronauts by stocking the required drugs in advance. Long space missions require novel approach to producing pharmacticals on short notice from pre-defined, limited raw materials.
Synthetic minimal cells provide just the right kind of programmable bioreactors that can be used to manufacture small molecule and biological drugs in short amount of time. Synthetic cells can be lyophilised and stored for a long time, and the programmability of the synthetic cell genetic circuits means almost every kind of chemical transition can be coded with the appropriate enzyme, and new enzymatic pathways for manufacturing drugs can be created on demand.
Our research focuses on designing mission-oriented solutions for preparation and storage of synthetic cells for the purposes of small scale, fast and responsive manufacturing of pharmaceuticals.
Synthetic biology
Most of the efforts of biological research, both in studying healthy organisms as well as disease states, is focused either on studying the whole live cell (or organism), or on studying isolated reactions between few purified and well defined components in vitro. Most biological processes are not isolated events of interaction between few components, but rather complex interconnected networks built of often multifunctional nodes (proteins acting on many targets). The in vitro studies give results that are less relevant to natural biology – since an experiment with a few purified components does not acknowledge the vast complexity of a natural system. On the other hand, live cell studies are notoriously hard to reproduce and interpret, due to the variability between live subjects, as well as due to the inherent complexity of biology (cross-talk between the studied process and other pathways, or background signal from unrelated processes is often present).
Synthetic minimal cells deliver a solution bridging the existing gap between in vitro and live cell research: use synthetic minimal cells to investigate multicomponent gene pathways, combining the advantages of in vitro systems with the relevancy and complexity approaching that of whole cell studies.
Reading and controlling cells is the core purpose of modern synthetic biology, and the overarching goal of all biomedical studies. Both studying mechanisms of most diseases, as well as investigating healthy cellular processes, is currently done as either in vitro or live cell experiments. In vitro research methods are easy to use, cheap and efficient way to obtain information about behavior of specific, well defined protein or nucleic acid complexes or single enzymes, or to characterize small molecule interactions between metabolites or drugs and their specific biological targets. However, since life is structured in complexes that involve many components organized in precise 3D assemblies, the in vitro experiments often only deliver information about small snapshot of this complex, natural system. Studies of live cells allow to obtain truly biologically relevant information about complex pathways, but at significantly higher cost, and with results that are harder to interpret and often less reproducible. The variability between live cell subjects and the underlying intricacy of interconnected biological networks constantly interacting with each other makes signal measured in live cell experiments often more noisy and the experiment itself difficult to design.
Synthetic minimal cells offer a platform that allows studying complex genetic pathways, while keeping the complexity of the system at a level that still allows us understanding fully what the system contains and how to engineer it. Our research focuses on building tools for general use in many areas of synthetic biology, as well as studying some specific cases of complex biological processes, both healthy and diseased, that are not accessible by studying natural complex cells.
Automation
We are developing solutions for automating and scaling thropought of synthetic cell epxeriments. Microfluidic rigs help us work faster, iterating more genetic circuit designs with automated liposome formation and streamlined detection of protein expression.
Our first rig recently started producing droplets with fluorescent RNA.
Astrobiology
Astrobiology is studying past, present and future of the life on the Universe. Synthetic cells are the perfect tool to answer many of the questions typically asked by astrobiologists, including engineering synthetic cells to resemble earliest terrestrial life forms for studying of the origins of life on Earth, engineering synthetic cell systems to explore possibilities of biochemistries yielding life on other planets, and exploring possible future evolution scenarios for various life forms. Using synthetic cells we can ask many questions about “weird life” events and alternative life forms, questions that would be nearly impossible to answer studying modern, complex terrestrial cells.
Our work is driven by the question of what is the multidimensional niche space for life, and what are the molecular mechanisms of these boundary physicochemical conditions of life? This leads naturally to the questions of how does terrestrial life responds to changes at those boundary conditions, and can synthetic biology be used to expand these? We aim at defining comprehensive matrix describing mechanisms in which environment restricts basic processes of biology, and developing framework for studying and designing living systems around those limitations.

Exploring the Informational Transitions Bridging Inorganic Chemistry and Minimal Life
Understanding what separates life from non-life is critical to solving some of the great outstanding questions in science, such as how life first emerged and how we might unambiguously detect life on other worlds. The transition from non-living to living matter has been notoriously difficult to quantify, limiting our ability to develop the necessary theory for understanding life or its universal properties. One of the most significant challenges is the complexity of the examples of life available to study, which represent the product of more than 3.5 billion years of evolution. Systems representing the intermediate stages of complexity between simple chemistry and modern biology are only now becoming accessible to study in the lab with advances in systems chemistry and minimal synthetic life, respectively. These have not yet been connected to explore the uncharacterized landscape between simple chemical systems and the much more complex biochemical architectures characteristic of life today. Our project is an unprecedented opportunity to systematically study the full pathway from non-life to life through a synthesis of bottom-up chemistry, top-down minimal biology and fundamental theory. We aim to build very simple systems from chemical and biological parts, and by studying varying stages of programmed complexity and autonomy, systematically evaluate the transitions in information through which these become more life-like, self-referential entities.
Thank you for support: Templeton Foundation.
This work is done in collaboration with Sara Walker, Paul Davies and Lee Cronin.
Build-a-Cell
Build-a-Cell is an international community of scientists and policymakers working on building synthetic cells. We facilitate collaborations between groups in different areas of synthetic cell research, we engage with scientists in other disciplines to promote use of synthetic cell tools, and we provide information for the general public.
Our lab participated in every one of the bi-annual Build-a-Cell workshops, and we are actively pursuing collaborations with many Build-a-Cell members.
Kate Adamala is the co-founder and leader of Build-a-Cell.
STAT magazine published nice overview of efforts to build artificial life, From chemicals to life: Scientists try to build cells from scratch.
Details, including list of member labs, working groups, workshops and all resources can be found at www.buildacell.org.
DIY Bio
The HTGAA course is based on the classic Fab-Lab “Hot to Make Almost Anything” fabrication class.
BioAcademy is the best resource to learn basisc of bioengineering, including specific applications of most cutting edge techniques. We run a synthetic cell module, including overwiew of the field of synthetic life and examples of synthetic cell based tools for biological engineering.
Excellent resource for beginning and advanced DIY biological engineers, biospaces and self-taught biohackers, as well as interesting supplement for anyone studying basics of modern biological engineering.
Synthetic cell tools developed by our lab
We have developed a toolbox of computational resources, custom devices and protocols for engineering synthetic cells.
|
|
|
|
Addgene deposits from out lab (plasmids and bacteria strains):
www.addgene.org/Kate_Adamala/
Coming soon:
Cyanoconstruct
Cellulator
Tools are free to use; CC BY-NC-SA 4.0.
![]()